Workflow example
Aryan Kamal, Judith Zaugg
15 December 2021
workflow.RmdAbstract
This workflow vignette shows how to use the GRaNPA package in a real-world example. Importantly, you will also learn in detail how to work with a GRaNPA object and what its main functions and properties are. The vignette will be continuously updated whenever new functionality becomes available or when we receive user feedback.
In the following example, you will use example data from the GRaNPA package. It contains an filtered eGRN object reconstructed using GRaNIE package for naive macrophage data and Dorothea GRN with confidence level of A. It also includes differential expression data from naive vs infected macrophages.
library(GRaNPA)## Loading required package: tidyverse## ── Attaching packages ─────────────────────────────────────── tidyverse 1.3.1 ──## ✓ ggplot2 3.3.3 ✓ purrr 0.3.4
## ✓ tibble 3.1.6 ✓ dplyr 1.0.7
## ✓ tidyr 1.1.4 ✓ stringr 1.4.0
## ✓ readr 1.4.0 ✓ forcats 0.5.1## ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
## x dplyr::filter() masks stats::filter()
## x dplyr::lag() masks stats::lag()## Loading required package: magrittr##
## Attaching package: 'magrittr'## The following object is masked from 'package:purrr':
##
## set_names## The following object is masked from 'package:tidyr':
##
## extractThe GRN object should always contain these two columns: ‘TF.name’, ‘gene.ENSEMBL’. additional column ‘weight’ is optional for GRNs containing weight.
GRN = GRaNPA::GRaNPA_eGRN_example
head(GRN)## TF.name TF_peak.r_bin TF_peak.r TF_peak.fdr TF_peak.fdr_orig
## 1 ALX1.0.B (-0.55,-0.5] -0.5459238 0.1501830 0.1501830
## 2 ASCL2.0.D [0.6,0.65) 0.6270437 0.1116630 0.1116630
## 3 BATF.0.A [0.55,0.6) 0.5695714 0.1835308 0.1835308
## 4 BATF.0.A [0.55,0.6) 0.5695714 0.1835308 0.1835308
## 5 CEBPB.0.A [0.6,0.65) 0.6491679 0.1768122 0.1768122
## 6 CEBPB.0.A [0.6,0.65) 0.6072994 0.1768122 0.1768122
## TF_peak.fdr_direction TF_peak.connectionType peak.ID
## 1 neg expression chr6:24757505-24758079
## 2 pos expression chr8:141401523-141401927
## 3 pos expression chrX:20322733-20322964
## 4 pos expression chrX:20322733-20322964
## 5 pos expression chr19:8540624-8541142
## 6 pos expression chr19:14589455-14589899
## peak.mean peak.median peak.CV peak.annotation peak.GC.perc peak.width
## 1 118.66751 120.71655 0.1655064 Distal Intergenic 0.4660870 575
## 2 66.39116 59.46970 0.3724172 Intron 0.6641975 405
## 3 48.01510 44.80189 0.3906979 Distal Intergenic 0.4094828 232
## 4 48.01510 44.80189 0.3906979 Distal Intergenic 0.4094828 232
## 5 42.32361 43.40715 0.4251483 Intron 0.4971098 519
## 6 25.04228 20.25975 0.5056223 Intron 0.4966292 445
## peak.GC.class peak_gene.distance peak_gene.r peak_gene.p_raw peak_gene.p_adj
## 1 (0.4,0.5] 16852 0.5187923 4.316523e-04 0.013773979
## 2 (0.6,0.7] 9528 0.4241387 5.121718e-03 0.070504465
## 3 (0.4,0.5] 198208 0.4689614 1.732104e-03 0.036145043
## 4 (0.4,0.5] 172822 0.5958414 3.138605e-05 0.001774220
## 5 (0.4,0.5] 177335 0.5604949 1.132026e-04 0.005011157
## 6 (0.4,0.5] 29218 0.4089397 7.167667e-03 0.087886371
## gene.ENSEMBL gene.mean gene.median gene.CV gene.chr gene.start
## 1 ENSG00000112312 637.322633 635.899554 0.3476213 chr6 24774931
## 2 ENSG00000184489 201.597187 190.336310 0.5461894 chr8 141391995
## 3 ENSG00000173674 2414.981186 2060.118304 0.3981834 chrX 20124525
## 4 ENSG00000177189 10490.978458 8712.855655 0.4106422 chrX 20149911
## 5 ENSG00000167772 142.499008 86.868304 0.9472575 chr19 8363289
## 6 ENSG00000131355 6.160821 4.830357 0.7883818 chr19 14619117
## gene.end gene.strand gene.type gene.name
## 1 24786099 + protein_coding GMNN
## 2 141432454 + protein_coding PTP4A3
## 3 20141838 - protein_coding EIF1AX
## 4 20267519 - protein_coding RPS6KA3
## 5 8374370 + protein_coding ANGPTL4
## 6 14690027 - protein_coding ADGRE3The DE dataset should always contain these three columns: ‘ENSEMBL’, ‘padj’ and ‘logFC’
DE = GRaNPA::GRaNPA_DE_example
head(DE)## ENSEMBL gene_name gene_synonym baseMean
## 1 ENSG00000136688 IL36G IL-1F9,IL-1H1,IL-1RP2,IL1E,IL1F9,IL1H1 4052.35038
## 2 ENSG00000007908 SELE CD62E,ELAM,ELAM1,ESEL 197.22505
## 3 ENSG00000108342 CSF3 C17orf33,G-CSF,GCSF,MGC45931 4231.99980
## 4 ENSG00000214107 MAGEB1 CT3.1,DAM10,MAGE-Xp,MAGEL1,MGC9322 15.19545
## 5 ENSG00000140955 ADAD2 FLJ00337,TENRL 46.32944
## 6 ENSG00000164400 CSF2 GM-CSF,GMCSF 1442.87905
## pvalue padj padj.beforeIHW logFC
## 1 0.000000e+00 0.000000e+00 0.000000e+00 12.73449
## 2 2.321540e-133 8.296570e-132 4.658950e-132 12.52533
## 3 0.000000e+00 0.000000e+00 0.000000e+00 12.40700
## 4 1.930085e-92 2.421056e-91 2.418017e-91 12.11311
## 5 2.130227e-74 2.096754e-73 2.093313e-73 12.06622
## 6 0.000000e+00 0.000000e+00 0.000000e+00 12.04483eGRN
This is the main function for GRaNPA. It will train a random forest based on provided GRN to predict the DE values. Here the DE values has been filtered by adjusted pvalue 0.2 and absolute log2 fold-change 2. Please follow (?getGRNConnections) for more information.
GRaNPA_result = GRaNPA::GRaNPA_main_function(DE_data = DE,
GRN_matrix_filtered = GRN,
DE_pvalue_th = 0.05,
logFC_th = 2,
num_run = 3,
num_run_CR = 2,
num_run_random = 3,
cores = 20,
importance = "permutation",
ML_type = "regression",
control = "cv",
train_part = 1)## INFO [2021-12-15 11:06:35] GRN Main evaluation function
## WARN [2021-12-15 11:06:35] In this version, the GRN should be filtered before
## WARN [2021-12-15 11:06:35] Both DE data and GRN should contain ENSEMBL ID for the Genes
## INFO [2021-12-15 11:06:35] Differential expression will be filltered by 0.05 pvalue and 2 logFC
## INFO [2021-12-15 11:06:35] Running GRaNPA for the Actual Network
## INFO [2021-12-15 11:06:35] Preparing the Gene-TF Matrix.
## INFO [2021-12-15 11:06:35] The matrix has been created.
## INFO [2021-12-15 11:06:35] Finished sucessfully. Execution time: 0.2 secs
## INFO [2021-12-15 11:06:35] Final data prepration finished.
## INFO [2021-12-15 11:06:35] The dimensions of final matrix are ( 270 , 115 ).
## INFO [2021-12-15 11:06:35] Finished sucessfully. Execution time: 0 secs
## INFO [2021-12-15 11:06:35] Actual network number : 1## Loading required package: lattice##
## Attaching package: 'caret'## The following object is masked from 'package:purrr':
##
## lift## INFO [2021-12-15 11:07:02] Maching learning model Finished
## INFO [2021-12-15 11:07:02] Finished sucessfully. Execution time: 27.4 secs
## INFO [2021-12-15 11:07:02] Actual network number : 2
## INFO [2021-12-15 11:07:28] Maching learning model Finished
## INFO [2021-12-15 11:07:28] Finished sucessfully. Execution time: 25.7 secs
## INFO [2021-12-15 11:07:28] Actual network number : 3
## INFO [2021-12-15 11:07:54] Maching learning model Finished
## INFO [2021-12-15 11:07:54] Finished sucessfully. Execution time: 25.8 secs
## INFO [2021-12-15 11:07:54] Actual network successfully finished
## INFO [2021-12-15 11:07:54] Finished sucessfully. Execution time: 1.3 mins
## INFO [2021-12-15 11:07:54] Running GRNeval for Random GRN
## INFO [2021-12-15 11:07:54] Random GRN number : 1
## INFO [2021-12-15 11:07:54] Preparing the Gene-TF Matrix.
## INFO [2021-12-15 11:07:54] The matrix has been created.
## INFO [2021-12-15 11:07:54] Finished sucessfully. Execution time: 0.3 secs
## INFO [2021-12-15 11:07:54] Final data prepration finished.
## INFO [2021-12-15 11:07:54] The dimensions of final matrix are ( 270 , 115 ).
## INFO [2021-12-15 11:07:54] Finished sucessfully. Execution time: 0 secs
## INFO [2021-12-15 11:08:26] Maching learning model Finished
## INFO [2021-12-15 11:08:26] Finished sucessfully. Execution time: 31.7 secs
## INFO [2021-12-15 11:08:26] Random GRN number : 2
## INFO [2021-12-15 11:08:26] Preparing the Gene-TF Matrix.
## INFO [2021-12-15 11:08:26] The matrix has been created.
## INFO [2021-12-15 11:08:26] Finished sucessfully. Execution time: 0.1 secs
## INFO [2021-12-15 11:08:26] Final data prepration finished.
## INFO [2021-12-15 11:08:26] The dimensions of final matrix are ( 270 , 115 ).
## INFO [2021-12-15 11:08:26] Finished sucessfully. Execution time: 0 secs
## INFO [2021-12-15 11:08:58] Maching learning model Finished
## INFO [2021-12-15 11:08:58] Finished sucessfully. Execution time: 31.7 secs
## INFO [2021-12-15 11:08:58] Random GRN number : 3
## INFO [2021-12-15 11:08:58] Preparing the Gene-TF Matrix.
## INFO [2021-12-15 11:08:58] The matrix has been created.
## INFO [2021-12-15 11:08:58] Finished sucessfully. Execution time: 0.2 secs
## INFO [2021-12-15 11:08:58] Final data prepration finished.
## INFO [2021-12-15 11:08:58] The dimensions of final matrix are ( 270 , 115 ).
## INFO [2021-12-15 11:08:58] Finished sucessfully. Execution time: 0 secs
## INFO [2021-12-15 11:09:32] Maching learning model Finished
## INFO [2021-12-15 11:09:32] Finished sucessfully. Execution time: 33.5 secs
## INFO [2021-12-15 11:09:32] Random GRN dataset successfully finished
## INFO [2021-12-15 11:09:32] Finished sucessfully. Execution time: 1.6 mins
## INFO [2021-12-15 11:09:32] Running for Complete Random
## INFO [2021-12-15 11:09:32] Complete Random samples : 1
## INFO [2021-12-15 11:09:32] Preparing the Gene-TF Matrix.
## INFO [2021-12-15 11:09:32] The matrix has been created.
## INFO [2021-12-15 11:09:32] Finished sucessfully. Execution time: 0.1 secs
## INFO [2021-12-15 11:09:32] Final data prepration finished.
## INFO [2021-12-15 11:09:32] The dimensions of final matrix are ( 270 , 115 ).
## INFO [2021-12-15 11:09:32] Finished sucessfully. Execution time: 0 secs
## INFO [2021-12-15 11:10:04] Maching learning model Finished
## INFO [2021-12-15 11:10:04] Finished sucessfully. Execution time: 32.4 secs
## INFO [2021-12-15 11:10:04] Complete Random samples : 2
## INFO [2021-12-15 11:10:04] Preparing the Gene-TF Matrix.
## INFO [2021-12-15 11:10:04] The matrix has been created.
## INFO [2021-12-15 11:10:04] Finished sucessfully. Execution time: 0.2 secs
## INFO [2021-12-15 11:10:04] Final data prepration finished.
## INFO [2021-12-15 11:10:04] The dimensions of final matrix are ( 270 , 115 ).
## INFO [2021-12-15 11:10:04] Finished sucessfully. Execution time: 0 secs
## INFO [2021-12-15 11:10:42] Maching learning model Finished
## INFO [2021-12-15 11:10:42] Finished sucessfully. Execution time: 37.6 secs
## INFO [2021-12-15 11:10:42] Complete Random dataset successfully finished
## INFO [2021-12-15 11:10:42] Finished sucessfully. Execution time: 1.2 mins
## INFO [2021-12-15 11:10:42] Finished sucessfully. Execution time: 4.1 mins
## [1] "finished!"After the function finished, you can see the result as a density plot:
GRaNPA::plot_GRaNPA_density(GRaNPA.object = GRaNPA_result, plot_name = "density.pdf", outputFolder = ".", width = 4, height = 4)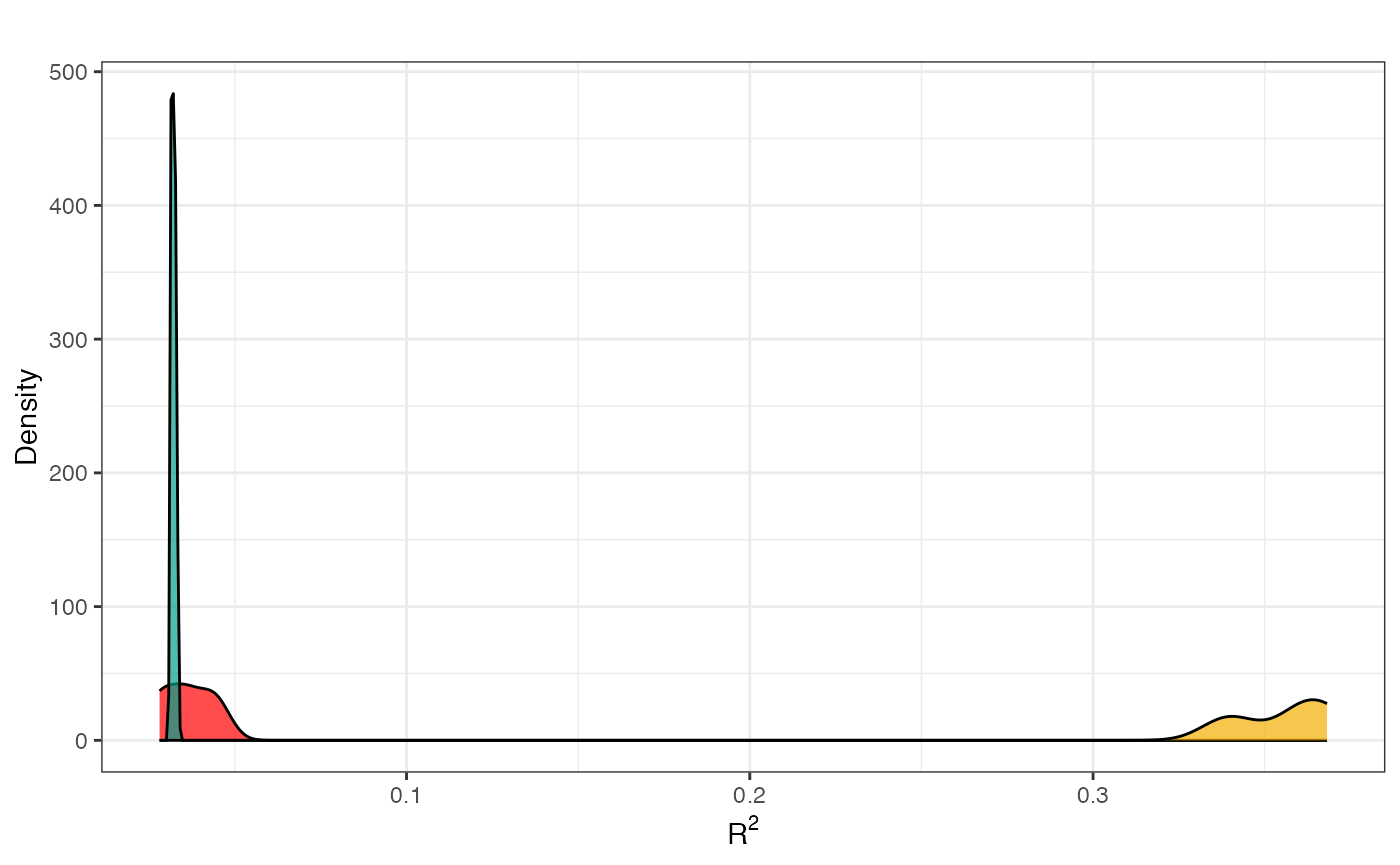
You can also see the relation between actual log fold change and predicted log fold change:
GRaNPA::plot_GRaNPA_scatter(GRaNPA.object = GRaNPA_result, plot_name = "scatter.pdf", output = ".", width = 4, height = 4) ## `geom_smooth()` using formula 'y ~ x'
## `geom_smooth()` using formula 'y ~ x'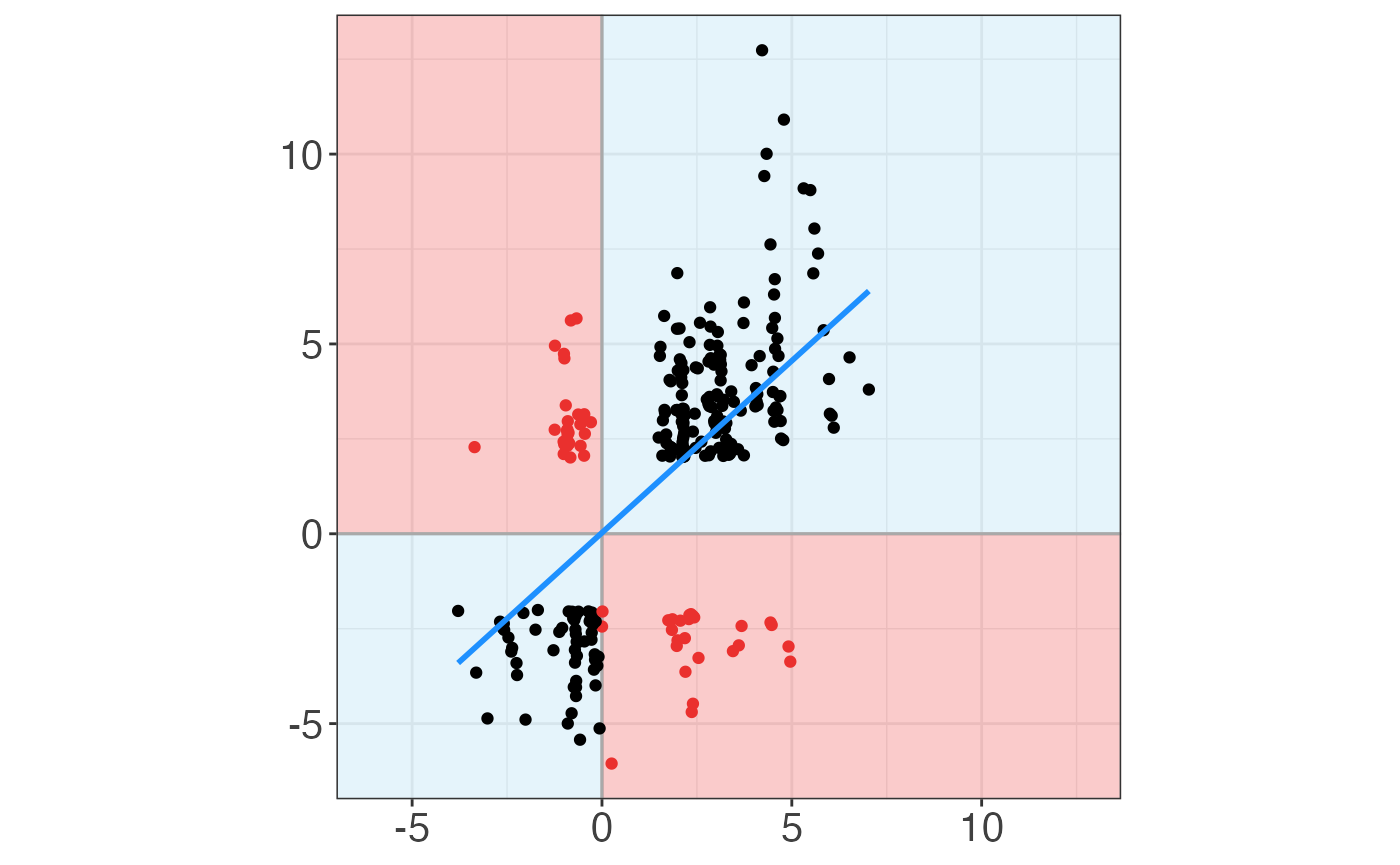
And using plot_GRaNPA_TF_imp function you can see important TFs:
GRaNPA::plot_GRaNPA_TF_imp(GRaNPA.object = GRaNPA_result, plot_name = "TF_imp.pdf", output = ".", width = 4, height = 4) 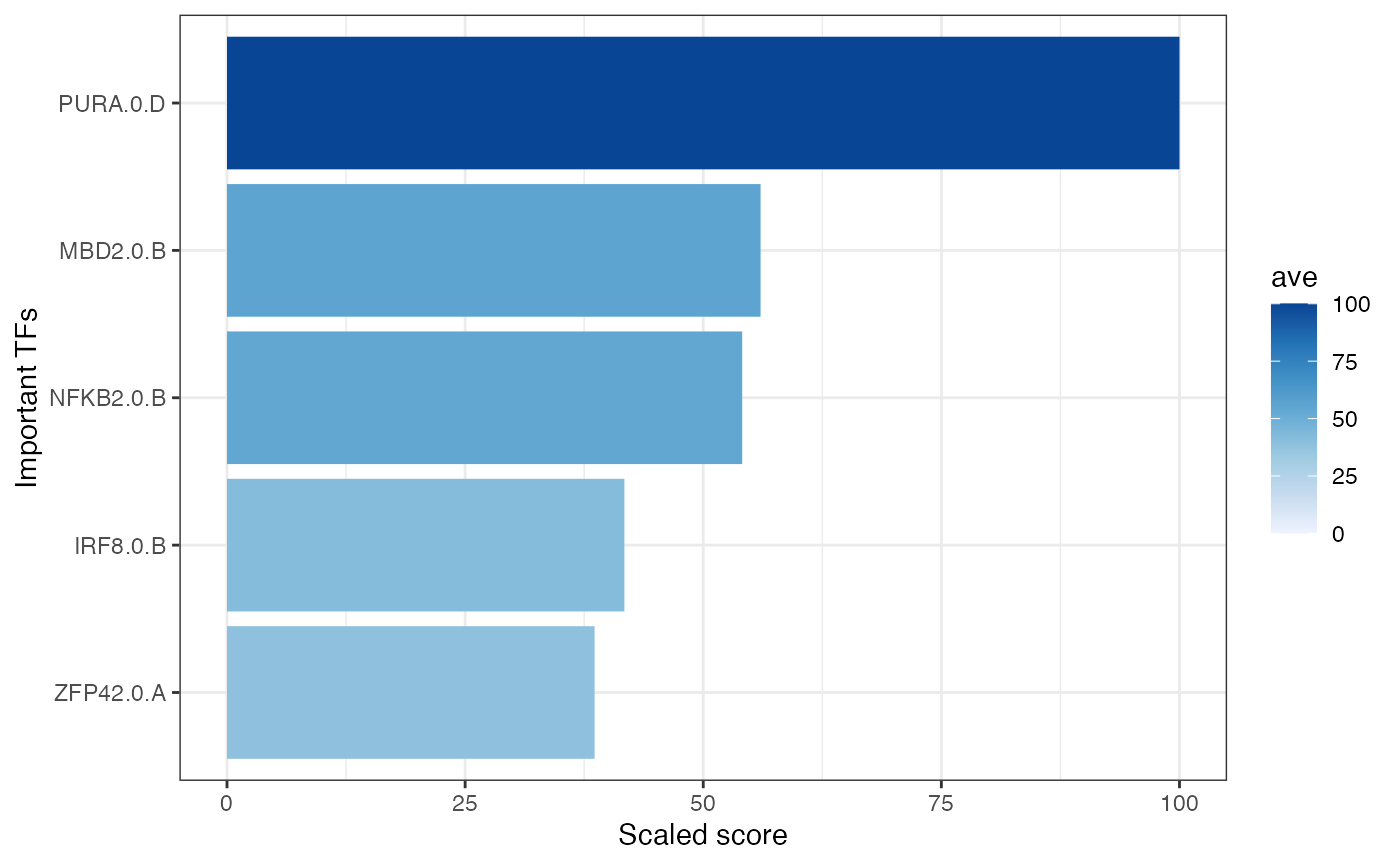
DoRothEA
Here, we also run GRaNPA on the Dorothea.
GRaNPA_result_dorothea = GRaNPA::GRaNPA_main_function(DE_data = DE,
GRN_matrix_filtered = GRaNPA::GRaNPA_Dorothea_A,
DE_pvalue_th = 0.2,
logFC_th = 2,
num_run = 3,
num_run_CR = 2,
num_run_random = 3,
cores = 20,
importance = "permutation",
ML_type = "regression",
control = "cv",
train_part = 1)## INFO [2021-12-15 11:10:47] GRN Main evaluation function
## WARN [2021-12-15 11:10:47] In this version, the GRN should be filtered before
## WARN [2021-12-15 11:10:47] Both DE data and GRN should contain ENSEMBL ID for the Genes
## INFO [2021-12-15 11:10:47] Differential expression will be filltered by 0.2 pvalue and 2 logFC
## INFO [2021-12-15 11:10:47] Running GRaNPA for the Actual Network
## INFO [2021-12-15 11:10:47] Preparing the Gene-TF Matrix.
## INFO [2021-12-15 11:10:48] The matrix has been created.
## INFO [2021-12-15 11:10:48] Finished sucessfully. Execution time: 1 secs
## INFO [2021-12-15 11:10:48] Final data prepration finished.
## INFO [2021-12-15 11:10:48] The dimensions of final matrix are ( 480 , 97 ).
## INFO [2021-12-15 11:10:48] Finished sucessfully. Execution time: 0.1 secs
## INFO [2021-12-15 11:10:49] Actual network number : 1
## INFO [2021-12-15 11:12:09] Maching learning model Finished
## INFO [2021-12-15 11:12:09] Finished sucessfully. Execution time: 1.3 mins
## INFO [2021-12-15 11:12:09] Actual network number : 2
## INFO [2021-12-15 11:13:20] Maching learning model Finished
## INFO [2021-12-15 11:13:20] Finished sucessfully. Execution time: 1.2 mins
## INFO [2021-12-15 11:13:20] Actual network number : 3
## INFO [2021-12-15 11:14:29] Maching learning model Finished
## INFO [2021-12-15 11:14:29] Finished sucessfully. Execution time: 1.1 mins
## INFO [2021-12-15 11:14:29] Actual network successfully finished
## INFO [2021-12-15 11:14:29] Finished sucessfully. Execution time: 3.7 mins
## INFO [2021-12-15 11:14:29] Running GRNeval for Random GRN
## INFO [2021-12-15 11:14:29] Random GRN number : 1
## INFO [2021-12-15 11:14:29] Preparing the Gene-TF Matrix.
## INFO [2021-12-15 11:14:29] The matrix has been created.
## INFO [2021-12-15 11:14:29] Finished sucessfully. Execution time: 0.3 secs
## INFO [2021-12-15 11:14:29] Final data prepration finished.
## INFO [2021-12-15 11:14:29] The dimensions of final matrix are ( 480 , 97 ).
## INFO [2021-12-15 11:14:29] Finished sucessfully. Execution time: 0 secs
## INFO [2021-12-15 11:15:56] Maching learning model Finished
## INFO [2021-12-15 11:15:56] Finished sucessfully. Execution time: 1.4 mins
## INFO [2021-12-15 11:15:56] Random GRN number : 2
## INFO [2021-12-15 11:15:56] Preparing the Gene-TF Matrix.
## INFO [2021-12-15 11:15:56] The matrix has been created.
## INFO [2021-12-15 11:15:56] Finished sucessfully. Execution time: 0.3 secs
## INFO [2021-12-15 11:15:56] Final data prepration finished.
## INFO [2021-12-15 11:15:56] The dimensions of final matrix are ( 480 , 97 ).
## INFO [2021-12-15 11:15:56] Finished sucessfully. Execution time: 0 secs
## INFO [2021-12-15 11:17:26] Maching learning model Finished
## INFO [2021-12-15 11:17:26] Finished sucessfully. Execution time: 1.5 mins
## INFO [2021-12-15 11:17:26] Random GRN number : 3
## INFO [2021-12-15 11:17:26] Preparing the Gene-TF Matrix.
## INFO [2021-12-15 11:17:26] The matrix has been created.
## INFO [2021-12-15 11:17:26] Finished sucessfully. Execution time: 0.3 secs
## INFO [2021-12-15 11:17:26] Final data prepration finished.
## INFO [2021-12-15 11:17:26] The dimensions of final matrix are ( 480 , 97 ).
## INFO [2021-12-15 11:17:26] Finished sucessfully. Execution time: 0 secs
## INFO [2021-12-15 11:18:53] Maching learning model Finished
## INFO [2021-12-15 11:18:53] Finished sucessfully. Execution time: 1.4 mins
## INFO [2021-12-15 11:18:53] Random GRN dataset successfully finished
## INFO [2021-12-15 11:18:53] Finished sucessfully. Execution time: 4.4 mins
## INFO [2021-12-15 11:18:53] Running for Complete Random
## INFO [2021-12-15 11:18:53] Complete Random samples : 1
## INFO [2021-12-15 11:18:53] Preparing the Gene-TF Matrix.
## INFO [2021-12-15 11:18:53] The matrix has been created.
## INFO [2021-12-15 11:18:53] Finished sucessfully. Execution time: 0.3 secs
## INFO [2021-12-15 11:18:53] Final data prepration finished.
## INFO [2021-12-15 11:18:53] The dimensions of final matrix are ( 480 , 97 ).
## INFO [2021-12-15 11:18:53] Finished sucessfully. Execution time: 0 secs
## INFO [2021-12-15 11:20:36] Maching learning model Finished
## INFO [2021-12-15 11:20:36] Finished sucessfully. Execution time: 1.7 mins
## INFO [2021-12-15 11:20:36] Complete Random samples : 2
## INFO [2021-12-15 11:20:36] Preparing the Gene-TF Matrix.
## INFO [2021-12-15 11:20:36] The matrix has been created.
## INFO [2021-12-15 11:20:36] Finished sucessfully. Execution time: 0.3 secs
## INFO [2021-12-15 11:20:36] Final data prepration finished.
## INFO [2021-12-15 11:20:36] The dimensions of final matrix are ( 480 , 97 ).
## INFO [2021-12-15 11:20:36] Finished sucessfully. Execution time: 0 secs
## INFO [2021-12-15 11:22:07] Maching learning model Finished
## INFO [2021-12-15 11:22:07] Finished sucessfully. Execution time: 1.5 mins
## INFO [2021-12-15 11:22:07] Complete Random dataset successfully finished
## INFO [2021-12-15 11:22:07] Finished sucessfully. Execution time: 3.2 mins
## INFO [2021-12-15 11:22:07] Finished sucessfully. Execution time: 11.3 mins
## [1] "finished!"Here is density plot for Dorothea.
GRaNPA::plot_GRaNPA_density(GRaNPA.object = GRaNPA_result_dorothea, plot_name = "density_dorothea.pdf", outputFolder = ".", width = 4, height = 4)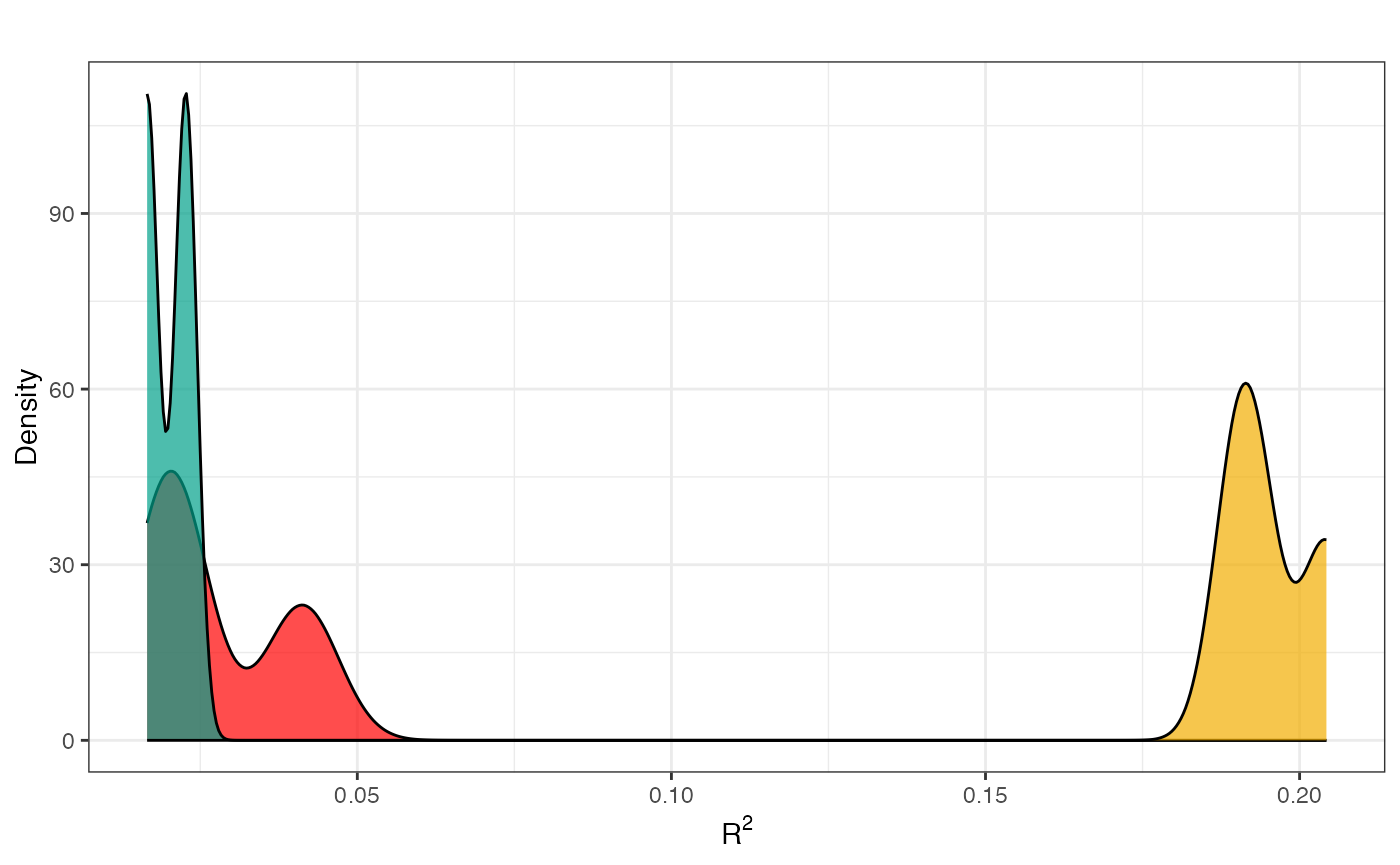
And using plot_GRaNPA_TF_imp function you can see important TFs:
GRaNPA::plot_GRaNPA_TF_imp(GRaNPA.object = GRaNPA_result_dorothea, plot_name = "TF_imp.pdf", output = ".", width = 4, height = 4) 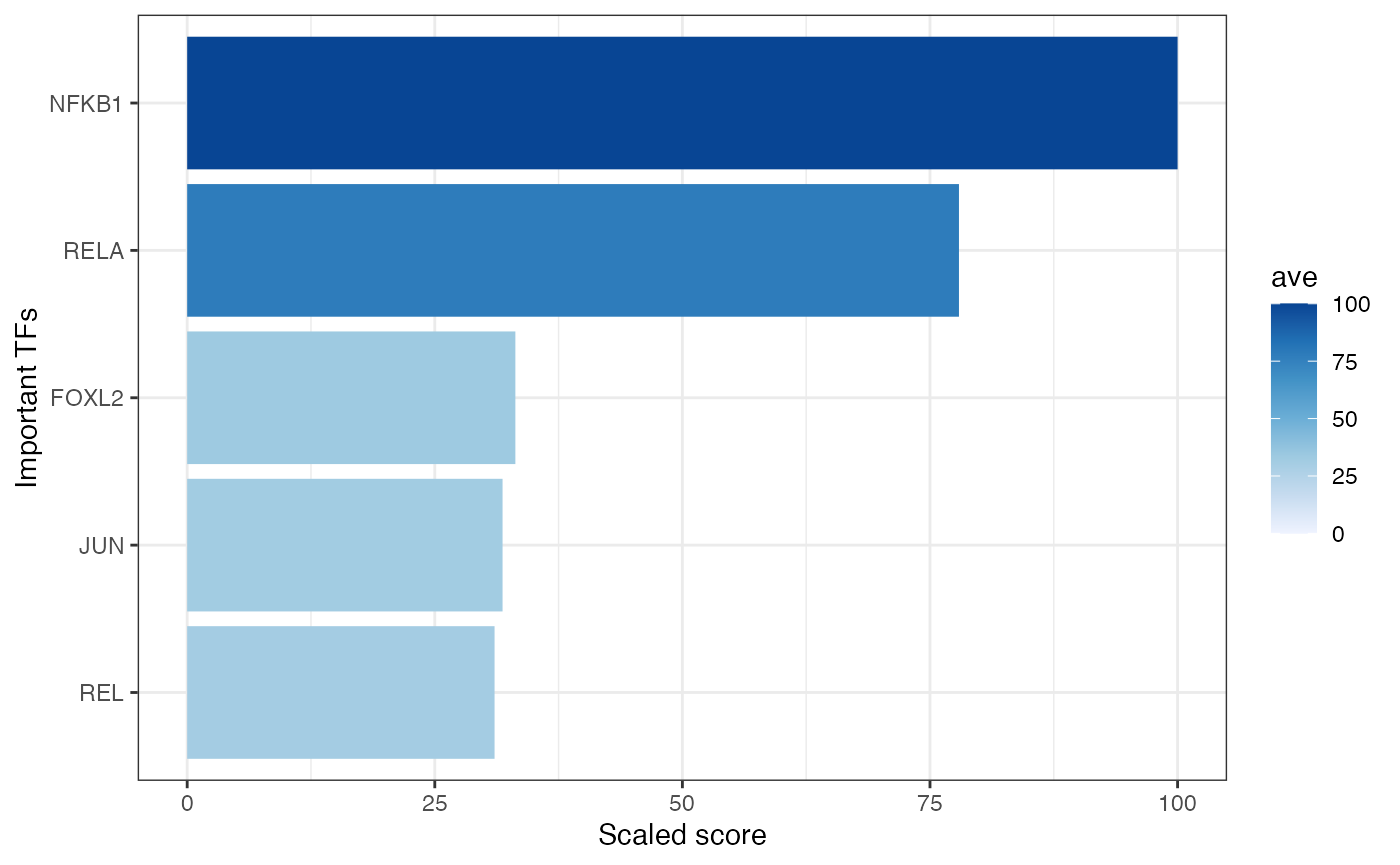
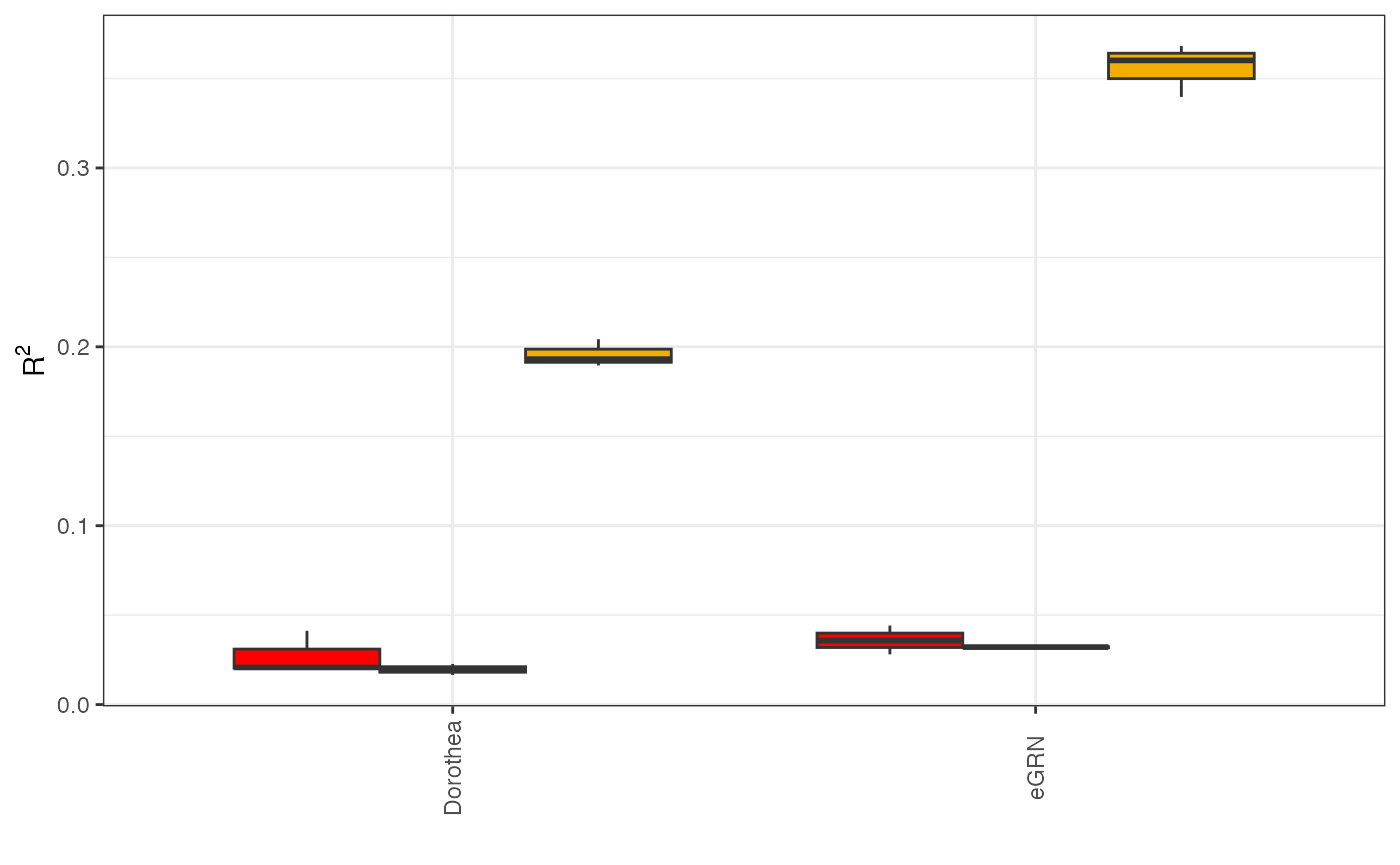
Network comparison
if you have several network and you want to compare them together you can use boxplot function
GRN_comp = list( network1 = GRaNPA_result_dorothea, network2 = GRaNPA_result)
GRaNPA::plot_GRaNPA_boxplot(GRaNPA.object_list = GRN_comp, name_list = c("Dorothea" , "eGRN"), plot_name = "comparison.pdf", output = "." , width = 8,height = 4)